Glia cell detection using FAIR's Detectron framework
- 4 minsGlial cell detection in human brain whole slide images
Deep learning in biology is getting increasingly popular due to the fact that bilologist have been using digital microscopy for decades now and their assumptions include tedious amounts of hard labour doing cell counting in whole slide images or annotating them almost pixel-by-pixel. A solution for this can be machine aided detection and annotation (or classification, region proposal and the list goes on and on).
Data
I’ve acquired some data from the University of Medicine in Budapest, Hungary from deciest Down-syndrome patients. Brain tissue have been thinly sliced and prepared for digital microscopy and the tissue was colorod with aldehid-dehydrogenase which is an enzyme for coloring glial cells in the brain.
In order to detect cell many hours of human labour was needed, so several biologists have counted cells on these huge images and annotated their locations in bounding boxes.
Annotation was done using Leica’s Aperio software.
Pre-processing
I wrote a small Python library that reads in a WSI (whole slide image) that is around ~8GB at a 40X magnification level and using ASAP I was able to read those in to memory by a predefined tile size tile-by-tile. The tiles were normalized and the annotation list was created from the Aperio xml output of annotations that containt the starting and ending coordinates of the glial cells along the longest diameter. From this I created bounding boxes and saves the starting coordinate of a cell and its width and height in a 4-dimensional vector.
Detectron
(FAIR)[https://research.fb.com/category/facebook-ai-research/] is one of the best AI Research facility in the world. They released the Detectron framework last year and since than it became popular and increasingly user friendly in a way that custom datasets and pre-trained models for transfer-learning can be easilly applied. It uses state of the art multi-object detection and segmentation networks that are based on the R-CNN, Faster R-CNN, Mask-RCNN and Pyramid Network architectures.
COCO
The above defined framework uses the COCO data definition in which annotations of pictures are stored in a special JSON format that includes the path of the pictures, their segmentation coordinates, their bonding box coordinates (of object). The architectures can be modified using the pre-defined yaml files. Learning rates, number of epochs, complete network architecture and testing, validating and training datasets can be set.
Metrics include intersection over union (IoU) at different thresholds called AP35, AP50, AP75 at different percentage thresholds. Mean average precision can be achieved by averaging over all categories. In my case one real category and one background category had to be defined since the framework requires at least two categories in order to work.
General
Multi-object detection is a hard task since there is no easy way to do it. All networks need to define a region proposal set up in order to find out at which regions to take into account when running the network. In Faster R-CNN this is done by a neural network as well but in R-CNN it was done by a selective search algorithm and the network needed to be run thousands of times for a single image. There have been improvements in recent years that all aimed at looking at images only once and producing multi-outputs (YOLO, SSD).
In basic classification and single object detection tasks the set up is usually the following:
- classification: a loss function is defined that penalizes miss-labeling of images, sequences, etc.
- single object detection: a simple regression problem to 4 real numbers: x, y, width, height coordinates
- contrary in multi-object detection when the network is asked to find objects on a never before seen image it needs to decide how many images there are and where they are and sometimes it also need to differentiate between same category obejcts or define different segmenetation masks for them
Results
I trained two pre-trained models with transfer learning using detectron and applied the result on the training images and never before seen test images. Here I only showcase the training set since the test set has not been yet validated.
Using a Faster R-CNN network with Feature Pyramid architecture:
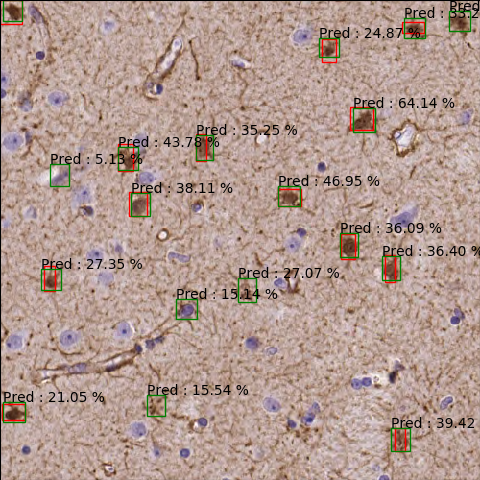
The percentages show accuracies. A cut has been applied at 3% minimum detection accuracy.
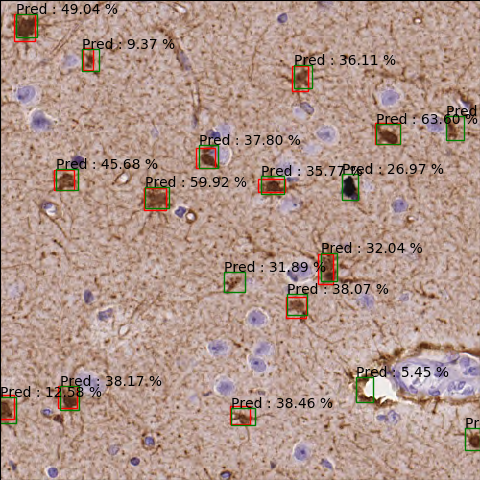
It can be seen that some cells are detected that have been not annotated (red) and some are miss-detected that have not been annotated but not glial cells.
Using a similar faster RCNN network:
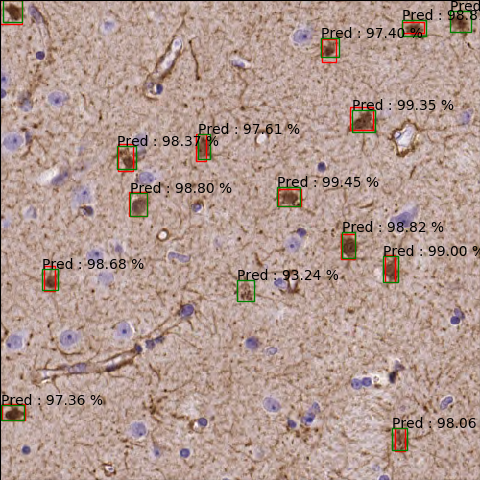
More accurate since predictions are made with higher percentages, the network is more confident:

My code can be found on GitHub. Please leave a star if you find my work intheresting.
