UNET architecture on multi-gpu for pathological image analysis
- 7 minsImage segmentation
Image segmentation is one of the many tasks of deep learning. One of the first architectures for image segmentation and multi-class detection was the UNET which uses a downsampling encoder and an upsampling decoder architecture with parameter sharing between different levels. The resulting neural network is trained with stochastic gradient descent with high momentum.
The original paper didn’t include batch normalization since it came out in just a few months before the proposal its proposal. They trained their model with a batch size of 1 image and the accompanying mask/s and their result was SOTA at the time. Since then many different implementations have been used that are more memory efficient and can be trained with bigger batch sizes.
My work
For my implementation I used PyTorch in order to get familiar with it. It seems much easier to use than TensorFlow but less straightforward than Keras, however, any model can be easilly extended to run on multiple GPUs and custom losses are easy to define. I will give a quick walk-through of my code here but will not go much into the details. Currently (2019-07-05) my code is not available online since I am using a not yet public dataset that I included in my repository, later I’ll include a GitHub link in this article.
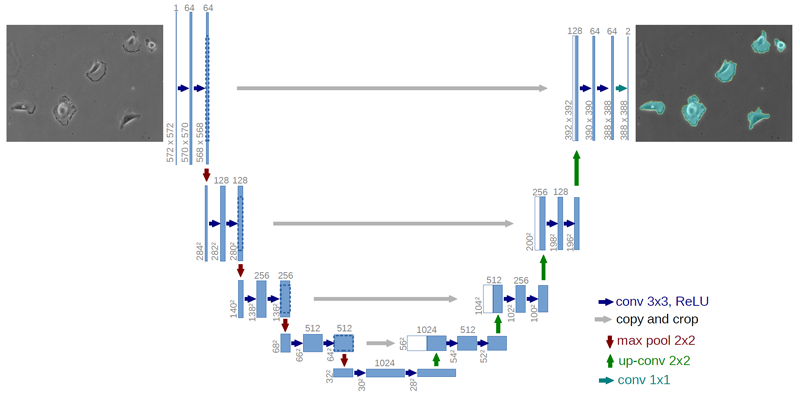
The architecture can be seen above, it consist of 4 convolutional blocks with max pooling (the encoder) and 4 convolutional blocks with transpose convolutional upsampling of data. Parameters are shared between all levels. The output can be a many layered image with a softmax activation if many classes are considered or a single layer mask with a sigmoid activation function to predict a binary mask. In my application I needed to predict binary masks only so I went with the sigmoid activation and binary cross entropy loss with the addition of the Jaccard-index that penalizes non-overlapping regions of predicted and ground truth masks.
\[J(A, B) = \frac{|A \cap B|}{|A| + |B| - |A \cap B|}\]The same can be read below, implemented in Python:
################################################
##### https://arxiv.org/pdf/1801.05746.pdf #####
################################################
class BCELossWithJaccard(Module):
def __init__(self):
super(BCELossWithJaccard, self).__init__()
self.bce = BCELoss()
def forward(self, x, y):
bce = self.bce(x, y)
xy = x*y
jaccard = torch.mean(xy / (x + y - xy))
return bce - torch.log(jaccard + 1e-12)
This expression is not completely correct since the predicted values are not binarized but I intended to make the system learn the correct binarization and also PyTorch didn’t provide a clear way of doing train time binarization.
I won’t include my whole implementation here but I used blocks like this in the encoder:
# Down forward pass
layer1 = self.layer_1_conv1(x)
layer1 = self.batch_norm_1_1(layer1)
layer1 = relu(layer1)
layer1 = self.layer_1_conv2(layer1)
layer1 = self.batch_norm_1_2(layer1)
layer1 = relu(layer1)
Blocks like this in the decoder:
# Up forward pass
up_layer_4 = interpolate(
layer5, scale_factor=2, mode='bilinear', align_corners=True)
up_layer_4 = self.upconv_4(up_layer_4)
up_layer_4 = torch.cat((layer4_crop, up_layer_4), dim=1)
up_layer_4 = self.uplayer_4_conv1(up_layer_4)
up_layer_4 = self.up_batch_norm_4_1(up_layer_4)
up_layer_4 = relu(up_layer_4)
up_layer_4 = self.uplayer_4_conv2(up_layer_4)
up_layer_4 = self.up_batch_norm_4_2(up_layer_4)
up_layer_4 = relu(up_layer_4)
Making the model run on multiple GPUs was pretty straigforward. All I needed to do was updating my pre-defined layers in a function to be data parallel:
def make_parallel(self):
# Layer 1
self.layer_1_conv1 = DataParallel(self.layer_1_conv1)
self.layer_1_conv2 = DataParallel(self.layer_1_conv2)
self.uplayer_1_conv1 = DataParallel(self.uplayer_1_conv1)
self.uplayer_1_conv2 = DataParallel(self.uplayer_1_conv2)
# ... code intentionally left out ... #
#######################################
Obviously my implementation is not perfect since some of my layers can’t be parallelized since I used the functional API for them and GPU usage is not equal, there is always one GPU that is used the most but it can run on any number of GPUs which is pretty amazing.
The library
Use of the library is pretty clean:
from patho import UNET
net = UNET()
net.make_parallel()
- the model can be initialized with any net and can be trained with binary cross entropy loss and binary cross entropy loss with jaccard index:
from patho import UNET, Model
net = UNET()
model = Model(net, lr=5e-3, with_jaccard=True)
- if the model was trained before a saved model is present in the
data/directory of the projectmodel.pt
from patho import UNET, Model
net = UNET()
model = Model(net, lr=5e-3, with_jaccard=True, load_model=True)
- for training one only needs to provide the data loader to the
.train(data_loader)function of the model
from patho import DataLoader
data_loader = DataLoader("patho/data/crc", "images",
"masks", batch_size=3).getInstance()
#
# ... code intentionally left out ...
#
model.train(data_loader)
Datasets
In this project I used two datasets. One of them is the original membrane dataset that is supposed to segment cells from context, so binary masks basically include borders between the cells. The other dataset is not yet public but contains colorectal tissue samples with the corresponding binary masks for cancerous regions. Permission for its usage can be accessed here.
I’ll present my results on these with this simple UNET architecture. For metric of detection goodness I use the Dice-score:
\[Dice-coefficient \quad := \quad \frac{2|X \cap Y|}{|X| + |Y|}\]Which is pretty similar to Jaccard-index but measures how much the ground truth masks correspond to the predicted masks.
It is self-explanatory that some kind of threshold must be applied to the predictions since not exact binary masks are provided. I was very conservative and used a 50%/70% threshold for calling a cell 1, thus cancer or cell tissue.
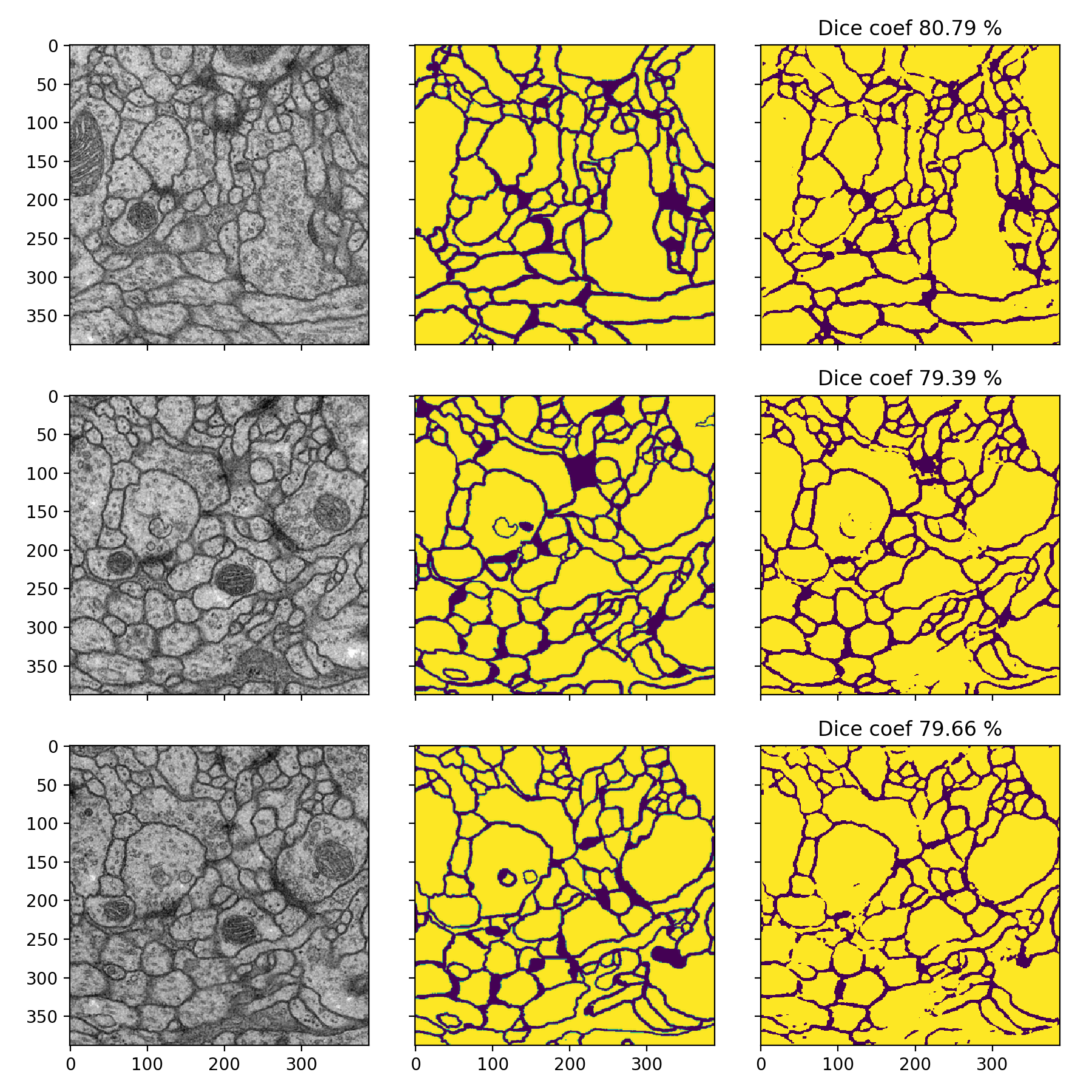
Membrane
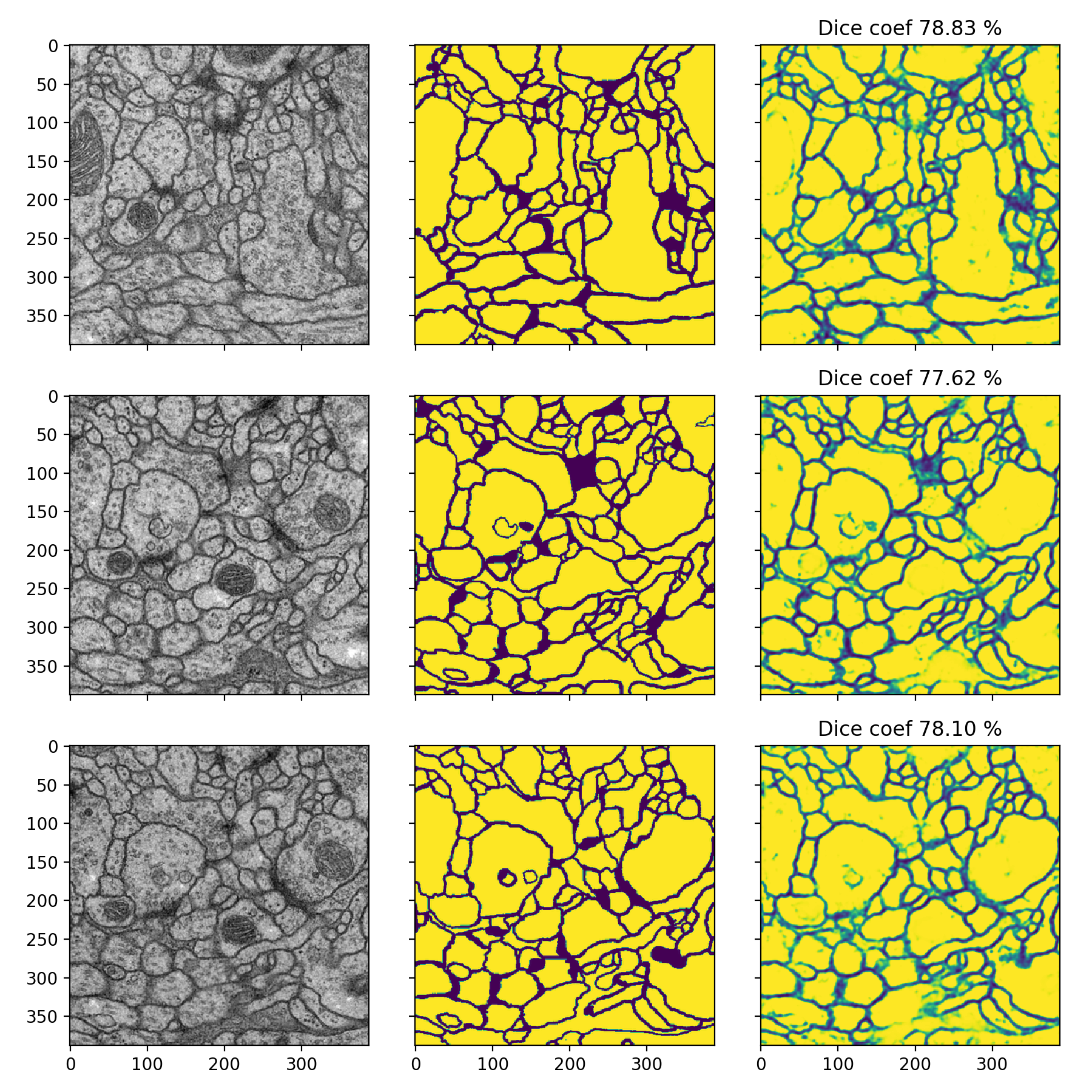
- Dice-coefficients are already pretty high but with the 50% thresholding they can be even better:
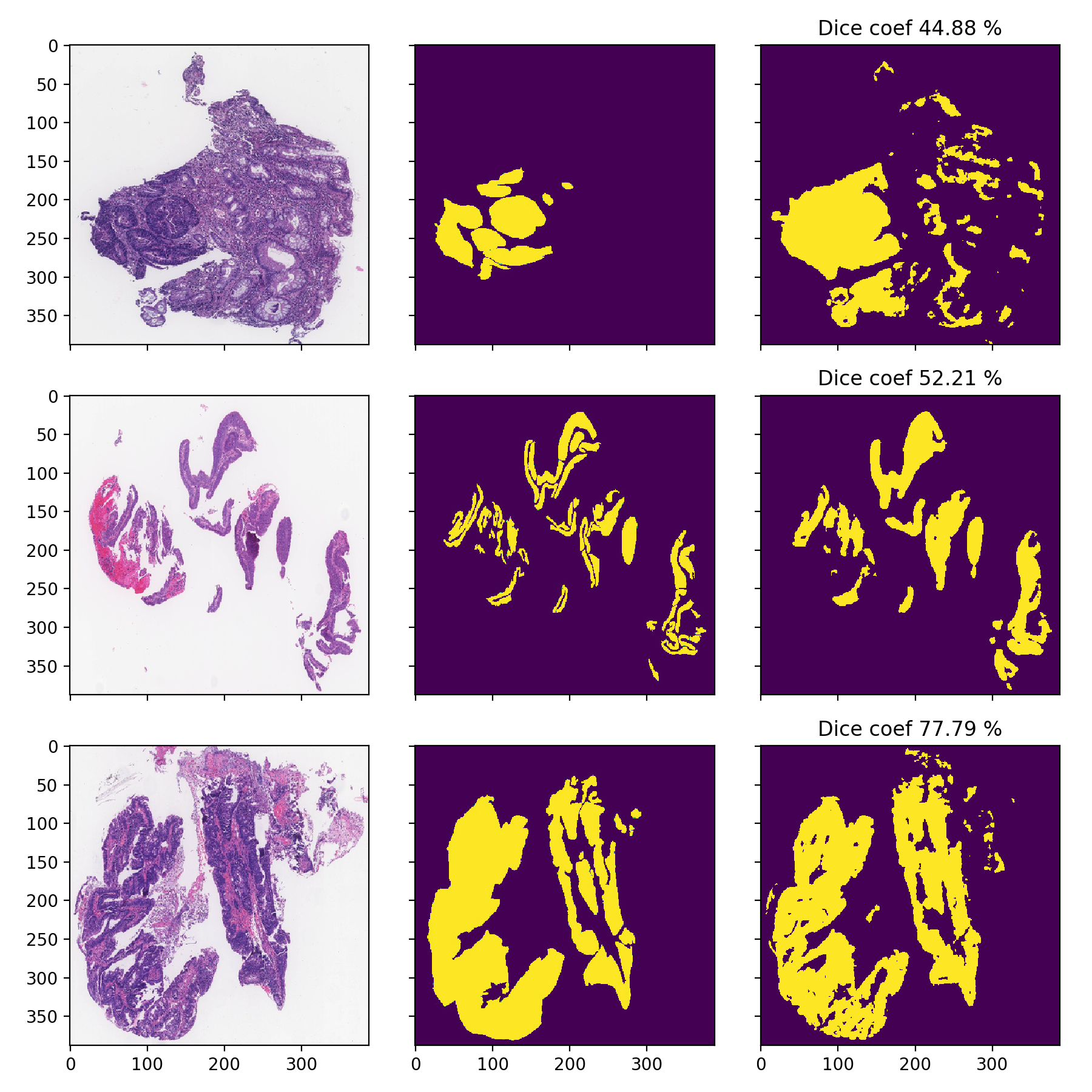
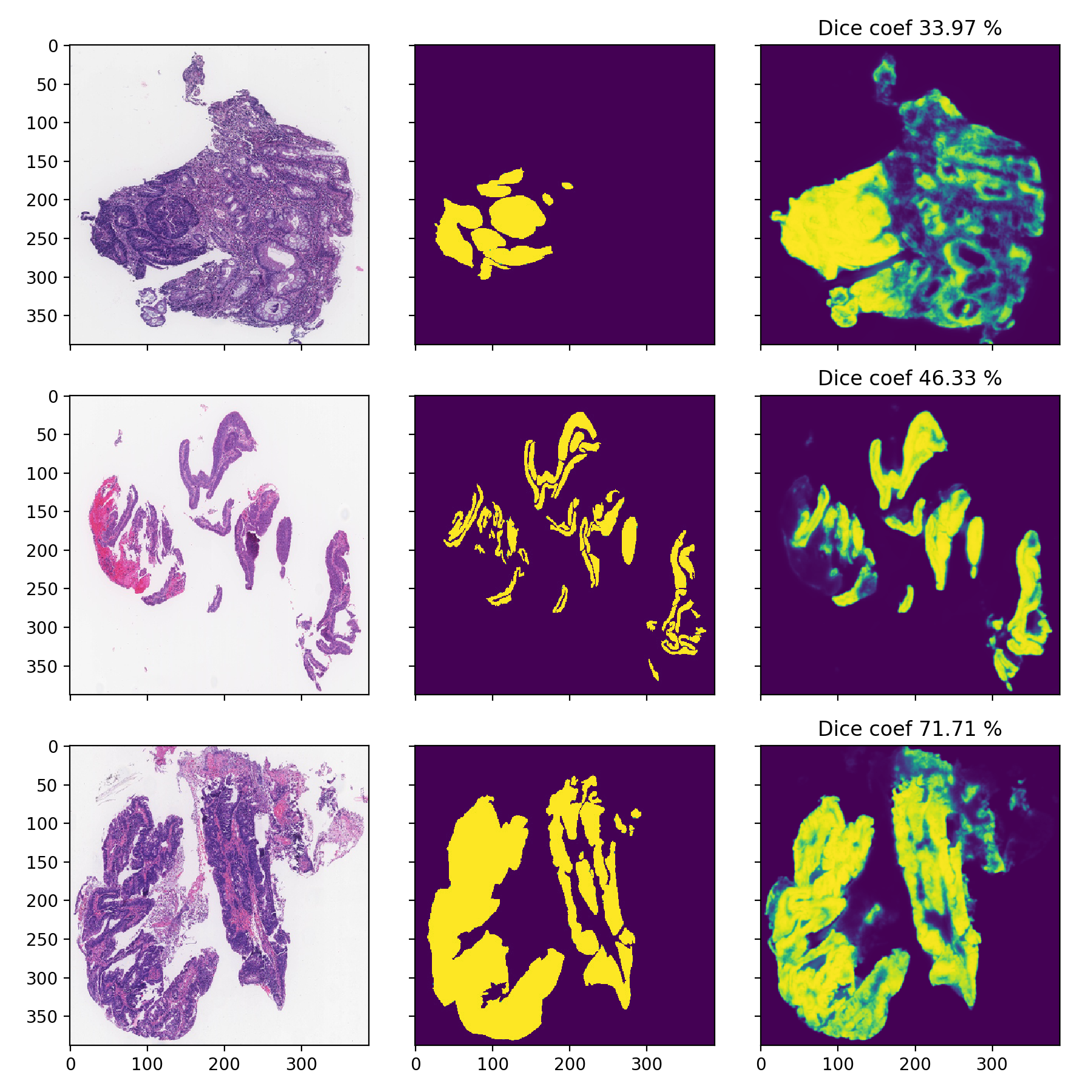
DigestPath
I am only showcasing these since these images are pretty low-resolution and can’t be used properly, the task is much harder as it can be seen from the significantly lower Dice-scores:
- Dice-coefficients are already pretty high but with the 70% thresholding they can be even better: