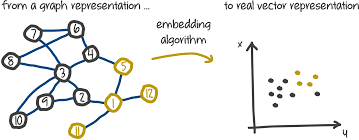
Graph representation learning - node embedding algorithms
- 10 minsGraph representation learning
Machine learning has succeded in many areas of science, numerous applications for computer vision has been seen but some might want to do the same for large graphs. Predict a label for each node or predict the probability of links to an other node, etc. This piece offers a small intro to how its done and lists some of the pioneering papers. The curated list is taken from here.
1. DeepWalk (2014)
DeepWalk proposes a way to embed vertices in a continouos vector space with respect to the topology of the graph. This way a classification algorithm could be run on the resulting vector embeddings or similarity measures defined for vectors could be experimented with.
The main idea is taken from NLP where word2vec was very successful. Here they build a corpus of vertex random walks from all nodes,f or limited length and for a fixed number of examples per node. This way nodes of large graphs can be embedded to a vector space via word2vec. The corpus is all the random walks where a sentence is one single random walk. Such as with word2vec a random matrix is initialized with the size of the vocabulary (number of nodes here) times the size of the desired embedding dimension. DeepWalk uses the skipgram model where the context is one single token and in a given window all the other vertices are being predicted.
2. GraRep (2015)
I don’t fully understand why the algorithm works but the algorithm itself is pretty straighforward. On the other hand it seems to me that for very large graphs this would not work due to the usage of the adjecency matrix and the SVD calculation. The algorithm is based on the k-step transition probability matrix that is calculated from the adjecency matrix and normalized by the node degrees.
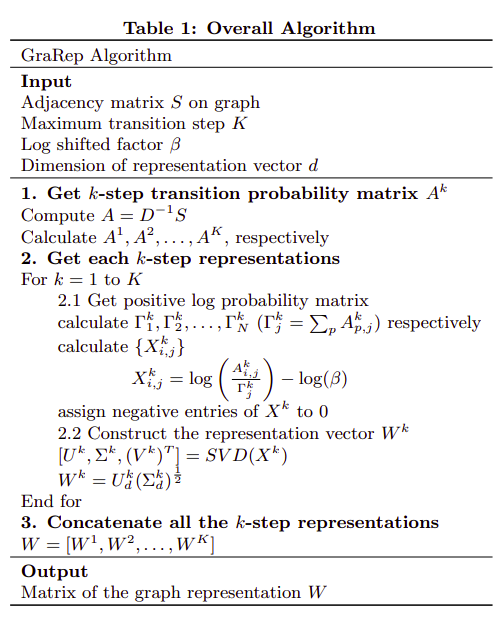
All the k-step representations are calculated and output by the algorithm.
The algoritmh itself is also based on an NLP idea of word embeddings.
3. Variational Graph Auto-encoders (2016)
They use graph convolutions so I will begin with that. It has been introduced by Kipf and Welling. They define the graph convolution as following (two layer version, for classification):
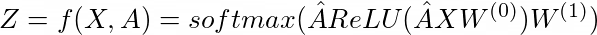
Where \(\hat{A}\) is the normalized adjecency matrix with self connections included:
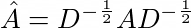
And \(W\) are the learnable weights. These are formulas derived by simplifications of the convolutional operation. It only includes first-neighbors and features a renormalization for computational stability.
In the VGAE paper they use this in order to derive meaningful latent representations using a matrix of latent embeddings and trying to restore \(\hat{A}\) by optimizing the ELBO. They use a non-variational method as well which is much simpler in equations:
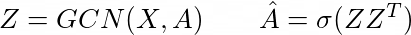
They perform better for link prediction than the DeepWalk algorithm (see. free lunch theorem).
4. Fast network embedding enhancement via high order proximity approximation (2017)
What they realised is that in all previous algorithms (node2vec, DeepWalk, GraRep, etc.) they all try to define proximity measures, such asd the kth order transition matrix:
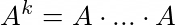
In all the previous works they use some kind of proximity measure as a higher order function of the adjecency matrix A (proven for DeepWalk as well). So in steps:
1. 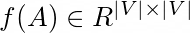 encodes Kth order proximity
encodes Kth order proximity
2. find f(A) = RC matrices that approximite it
They propose a method to update these matrices R, C such that they provide a higher order approximation g(A) by the simple rules:
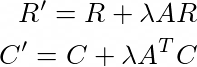
5. Poincaré embeddings for learning hierarchical representations (FAIR - 2017)
I do not understand this, but the main idea is that hyperbolic space is a continouos limit of the tree-like structure of hierarchical datasets and therefore it is a good bias on modelling such datasets. It is also scaleable via Riemann-optmization (???). It has ~500 citations and probably is pretty important but I think I need a matematician in order to tell me more about the matematical intuitions behind it.
6. Network embedding as matrix factorization (2018)
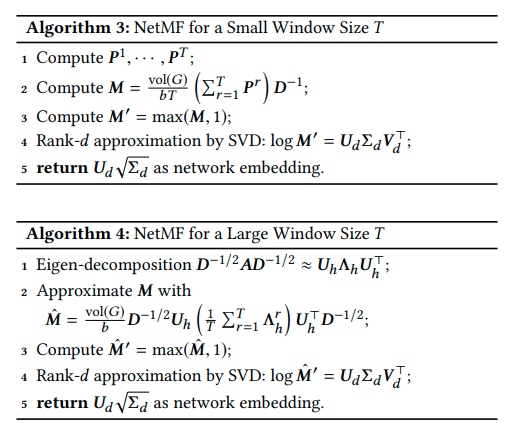
It sums up DeepWalk, PTE, LINE, node2vec algorithms in a single category and provides proofs and theorems in order to understand their similair nature as matrix factorization methods. Given this they prove that DeepWalk is probably the superior in theory out of all so they develop a new algorithm based on factorizing the DeepWalk derived matrix in order to achieve great embeddings. Here I provide the NetMF algorithm from the paper where the power of P are the normalized transition probabilities:
7. Adversarial network embedding (2017)
They build upon the DeepWalk algorithm and the shifted PPMI matrix X as input features for a generator network. Their work consists of three networks. Networks F() and G() take the features of a node from X as inputs and provide a low dimensional embedding for it. Based on the skip-gram negative sampling model they build the dataset with DeepWalk and create it in a tabular form:
| node i | node j | is context? |
|---|---|---|
| ‘2’ | ‘14’ | 1 |
| ‘2’ | ‘11’ | 0 |
| ‘2’ | ‘33’ | 0 |
| ‘2’ | ‘19’ | 1 |
| ‘2’ | ‘14’ | 1 |
The objective for, as they call it, inductive deep walk:
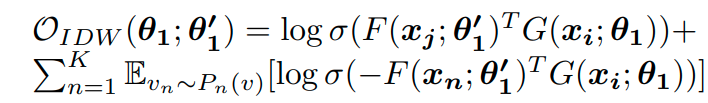
This loss term is only optimized for the parameters of F which produces the context embeddings and not the actual vertex embeddings. Alongside this term they try to optmize the embedding to be similar to samples from a prior distribution p(z) and play a minimax game between the generator network G() and the discriminator network D() which tries to separate generated embedding from ‘real’ samples. The discriminator and generator losses are the following:
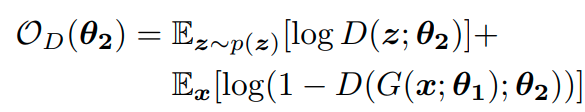
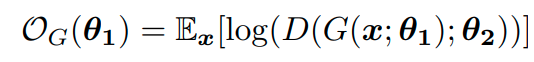
They don’t fully explain it but I imagine that they train all three networks jointly with the sum of the loss terms as the final objective and they produce on-par with DeepWalk and sometimes surpassing it by a small margin.
8. A general view for network embeddings as matrix factorization (2019)
It is the best paper so far describing and uniteing all the methods seen so far. Basically sums up network embedding tewchniques as matrix factorization and proposes a new method and also leaves room for other algorithms to be developed in this framework. Although has its limitations due to matrix inversion and the applciation of SVD as all the others before. So it cannot be used for million node graphs. The general form is basically the need to factorize:
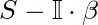
Where S is the user defined similarity matrix of vertices/nodes present, which can be some power of the adjecency matrix or the Laplacian or any other user-defined measure for node similarity. Thew objective to achieve a good embedding of the nodes can be generalized to making similair nodes close to each other while keeping similar embedding close to each other and also by keeping not similar nodes distant while doing so with the embeddings. In order to do this they state that increasing and decreasing functions are needed respectively:
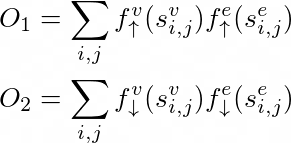
The total loss is the sum of the first and the weighted second term. They also provide specific functions to show how other methods can be included in this framework.
TAKEAWAY: GRA-beta method (that they present - gra: similarity, beta: tuning the parameter of S) is the best link predictor so far.
9. Network embedding as sparse matrix factorization (2019)
This is an iteration over the NetMF algorithm by the same authors. In this paper they provide a solution for embedding large network with tens of millions of nodes in reasonable time. I did not read the paper fully since it is very dry.
TAKEAWAYS: It seems that this method was they best in 2019 to embed very large networks into low dimensional vector spaces and the only other method capable of this was LINE with much lowe downstream accuracies. However if speed is a problem, LINE should be used since this algorithm is still pretty slow (24 hours on a many core CPU for ~100 million nodes).
10. vGraph: a generative model for joint community detection and node representation learning (2019)
This article is my next favorite after DeepWalk since it employs a generative model based on community and aims to predict neighboring vertices. The input for the learning algorithm is the edges (c, w) - node pairs, and the goal of the algorithm is to detect communities in an unsupervised manner from graph structure only.
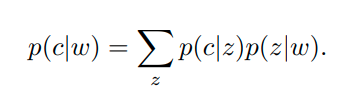
Here z is the variable for representing the community while c, w are representing nodes. An edge is generated via community sampling of the node and node sampling of the community to sum up what this article says. We should parametrize these probabilities with embeddings from nodes, context nodes and communities.
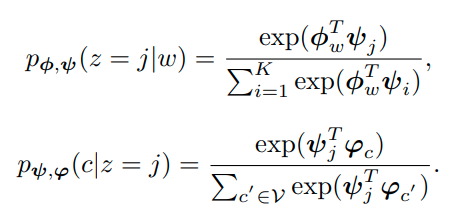
The latter cannot be computed efficiently this way for very large graphs so we would need to apply negative sampling that approximates this appropiately:
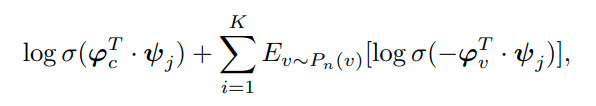
We approximate the community sampling conditional distribution with:
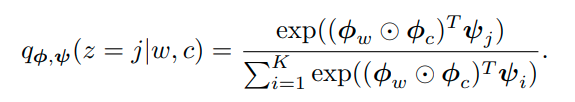
They optimize the expectation lower bound (ELBO) as they try to reconstruct the edge with the generative process and morph the approximate posterior into the prior with the KL-divergence term:
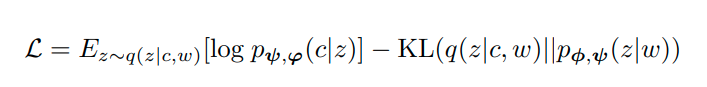
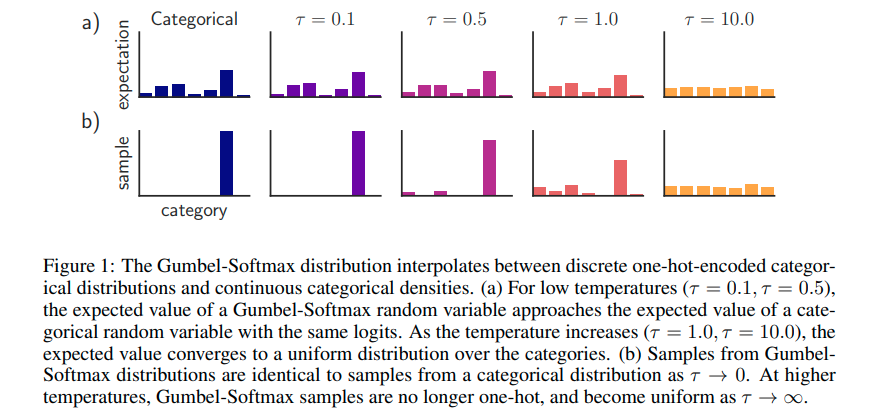
They also use a regulrization technique for smoother communities but that is not important regarding the theoretical foundation. One thing to mention is that they use the Gumbel-softmax-reparametrization which a reparametrization trick for categorical latent variables such as z. The main idea behind it is to sample a random uniform value and make the probability mass function concentrated on one single class. In practice this means that they take the community embedding and resample it with the Gumbel-softmax distribution on the community categories. Here is a good figure from the original paper:
11. GraphZoom (2020)
GraphZoom is a 2020 ICLR paper. It is basically defines a new way to add node features to topological structure via graph fusion. They build a kNN graph on the node features and weigh it by the cosine similarity between the node features. This matrix is then added with a weighing factor to the adjecency matrix of the original graph. This introduces new weights and can reweigh existing ones as well.
I just skimmed the rest of the paper. They use a high-pass filter on the eigenvalues of the Laplacian of this fused adjecency matrix and reduce the dimensonality that way. They impose projection matrices this way and therefore they can run graph embedding algorithms for very large graphs as well. Basically any graph embedding methodology can be used here, but they propose their own obviously.
Conclusion
There are some essential algorithms that I did not cover here, such as LINE or PTE, but the main idea behind it has been covered in some of the generalizing articles above. I am currently digging into graph neural networks and expect a new piece sometime in the next two weeks.
